| CPC C07H 19/00 (2013.01) [C07H 19/06 (2013.01); C07H 19/16 (2013.01); C12Q 1/42 (2013.01); C12Q 1/48 (2013.01); C12Q 1/6869 (2013.01); C12Q 1/6883 (2013.01); G01N 33/48721 (2013.01); B82Y 5/00 (2013.01); C12Q 2600/158 (2013.01); G01N 2333/9125 (2013.01)] | 20 Claims |
|
1. A method for nucleic acid sequencing, the method comprising providing an array of individually addressable sites, each site having a nanopore coupled to a nucleic acid polymerase, wherein the polymerase is attached to said nanopore within 0.7 nm of the entrance to said nanopore, and, at a given site of said array, polymerizing tagged nucleotides with the polymerase, wherein each tagged nucleotide comprises a tag capable of being cleaved in a nucleotide polymerization event and detected with the aid of the nanopore, wherein the tag is released and detected by the nanopore at said given site, wherein the nanopore is inserted into solvent-free planar lipid bilayer membranes, wherein the nucleotide sequence of a single-stranded DNA is determined, and comprises:
a) contacting the single-stranded DNA, wherein the single-stranded DNA is in an electrolyte solution and wherein the single-stranded DNA has a primer hybridized to a portion thereof, with:
(I) a nanopore of said array of individually addressable sites,
wherein the nanopore is an individually addressable nanopore in a membrane,
and
(II) at least four of said tagged nucleotides,
wherein each tagged nucleotide is a deoxyribonucleotide polyphosphate (dNPP) analogue,
under conditions permitting the polymerase to catalyze incorporation of one of the dNPP analogues into the primer if it is complementary to the nucleotide residue of the single-stranded DNA which is immediately 5′ to a nucleotide residue of the single-stranded DNA hybridized to the 3′ terminal nucleotide residue of the primer, so as to form a DNA extension product, wherein each of the four dNPP analogues has the structure:
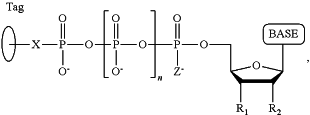 wherein the base is adenine, guanine, cytosine, thymine or uracil, or a derivative of one or more of these bases, wherein R1 is OH, wherein R2 is H or OH, wherein X is a linker and may comprise O, NH, S, or CH2, wherein n is 1, 2, 3, or 4, wherein Z is O, S, or BH3,
wherein the tag comprises an oligonucleotide which comprises nucleotides having natural bases and (i) at least one abasic site or (ii) at least one nucleotide having a modified base, with the proviso that:
(i) the type of base on each dNPP analogue is different from the type of base on each of the other dNPP analogues; and
(ii) the type of tag on each dNPP analogue is different from the type of tag on each of the other dNPP analogues,
b) polymerizing a complementary dNPP analogue from step a) into the primer to form the DNA extension product, wherein during polymerization a tag is released and detected by a nanopore by applying a voltage across the membrane and measuring an electronic change across the nanopore resulting from the tag translocating through the nanopore,
wherein the electronic change is different for each different type of tag, thereby permitting identifying of the nucleotide residue in the single-stranded DNA complementary to the incorporated dNPP analogue; and
c) repeatedly performing steps (a) and (b) for each nucleotide residue of the single-stranded DNA being sequenced,
thereby determining the nucleotide sequence of the single-stranded DNA.
|