| CPC G16B 45/00 (2019.02) [G06V 10/751 (2022.01); G06V 10/82 (2022.01); G06V 20/698 (2022.01); G16B 20/00 (2019.02); G16B 40/00 (2019.02)] | 20 Claims |
|
1. A computer-implemented method comprising:
receiving a plurality of phenomic images corresponding to a plurality of cell perturbations;
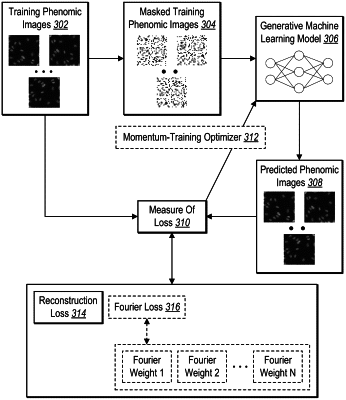
generating, utilizing a masked autoencoder generative model, a plurality of phenomic image autoencoder embeddings for the plurality of cell perturbations from the plurality of phenomic images, wherein the masked autoencoder generative model is trained to generate predicted phenomic images from masked training phenomic images corresponding to training phenomic images;
generating perturbation comparisons utilizing the plurality of phenomic image autoencoder embeddings; and
providing, for display within a graphical user interface, the perturbation comparisons for the plurality of phenomic image autoencoder embeddings.
|