| CPC G16B 30/00 (2019.02) [G06F 16/2255 (2019.01); G06F 16/2365 (2019.01)] | 27 Claims |
|
1. A method of filtering out erroneous sequence reads, the method comprising:
at a computer system having one or more processors, and memory storing one or more programs for execution by the one or more processors:
(A) obtaining a plurality of sequence reads, in electronic form, from a multiplexed sequencing reaction, wherein
the plurality of sequence reads comprises 10,000 sequence reads;
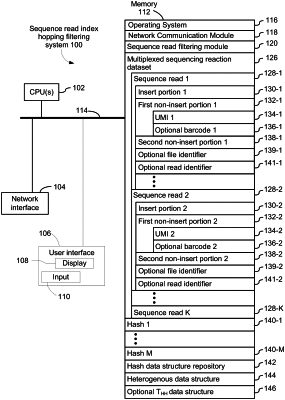
each respective sequence read in the plurality of sequence reads comprises a corresponding insert portion, a corresponding first non-insert portion consisting of N nucleotide positions, and a corresponding second non-insert portion consisting of M nucleotide positions,
N and M are positive integers,
each corresponding first non-insert portion represents an identifier of a unique molecule,
each corresponding second non-insert portion comprises a sample index,
the plurality of sequence reads includes one or more first sequence reads from a first sample that each have a first value for the sample index,
the plurality of sequence reads includes one or more second sequence reads from a second sample that each have a second value for the sample index;
(B) performing a uniqueness test by:
(i) obtaining a table THH that comprises the corresponding first and second non-insert portions of each sequence read in the plurality of sequence reads, lexicographically sorted on the first non-insert portion,
(ii) identifying each respective first non-insert portion represented in THH for which a predetermined second threshold number of unique second non-coding portion pairs can be made from pairs of sequence reads in Tim having the respective first non-insert portion, wherein each second non-coding portion pair in the threshold number of unique second non-coding portion pairs comprises an independent pair of different second non-coding portion values, and
(iii) removing from the plurality of sequence reads each sequence read in the plurality of sequence reads having an identified first non-insert portion;
(C) for each respective hash in a plurality of hashes, forming a corresponding hash data structure, wherein the corresponding hash data structure includes a representation of each respective sequence read, in the plurality of sequence reads, and comprises a hash value, of length L nucleotide positions, wherein L is less than N, formed by hashing at least the corresponding first non-insert portion of the respective sequence read in accordance with the respective hash, thereby forming a plurality of hash data structures;
(D) creating a heterogenous data structure comprising a plurality of entries by identifying a plurality of unique sequence read pairs in the plurality of hash data structures in greater than linear time, wherein
each respective sequence read pair in the plurality of unique sequence read pairs includes a corresponding first sequence read and a corresponding second sequence read sharing a common hash value and having different second values for the sample index, and
each corresponding entry in the heterogeneous data structure comprises a respective unique sequence read pair in the plurality of unique sequence read pairs, wherein the corresponding entry includes at least the corresponding first and second non-insert portions of the corresponding first sequence read and the corresponding first and second non-insert portions of the corresponding second sequence read of the respective unique sequence read pair;
(E) identifying a set of sequence reads using the heterogeneous data structure, wherein each sequence read in the set of sequence reads has a corresponding first non-insert portion value that appears more than a predetermined first threshold number of times in the heterogeneous data structure; and
(F) removing the set of sequence reads from the plurality of sequence reads thereby filtering out erroneous sequence reads.
|