| CPC G01N 21/6458 (2013.01) [G01N 21/6428 (2013.01); G01N 33/5005 (2013.01); G01N 33/582 (2013.01); G01N 33/6803 (2013.01); G01N 2021/6439 (2013.01); G01N 2201/12 (2013.01)] | 23 Claims |
|
1. A method of defining molecular architecture of a fluorophore labelled structure in a biological sample, the method comprising:
capturing a series of single molecule localization microscopy (SMLM) optical images of said structure, wherein said structure comprises a protein labelled with a fluorophore;
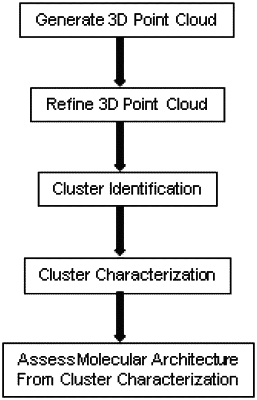
mapping a three-dimensional (3D) location of each single emission event in a plurality of single emission events from the series of SMLM optical images to create a 3D point cloud;
refining the 3D point cloud by merging two or more single emission events;
removing one or more single emission events that correspond to randomly generated points;
completing a multi-threshold network analysis to identify clusters within the refined 3D point cloud;
identifying non-biological network nano-clusters resulting from multiple emissions from a single fluorophore in the refined 3D point cloud, wherein multiple emissions from the single fluorophore are two or more emission events within 20 nm;
removing the non-biological network nano-clusters from the refined 3D point cloud; and
extracting features for each cluster at a threshold of 60 nm-140 nm to obtain size, shape and topology of each cluster and thereby define the molecular architecture of the fluorophore labelled structure;
wherein the extracting features for each cluster uses machine learning;
wherein non-biological networks are distinguished from biological networks in the 3D point cloud by performing a multi-scale network analysis of the 3D point cloud and determining a network degree distribution for each proximity threshold; and
wherein the refining of the 3D point cloud optionally comprises iterative steps.
|