| CPC C12Q 1/6816 (2013.01) [C12Q 1/6806 (2013.01); C12Q 1/6813 (2013.01); C12Q 1/6869 (2013.01)] | 16 Claims |
|
1. A method of improving performance of molecular inversion probe capture reactions, the method comprising:
fragmenting nucleic acid into a plurality of fragments by heating a buffer comprising the nucleic acid to about 65 degrees C. for at least about 16 hours to shift pH of the buffer to between about 6 and about 6.5 to hereby initiate acid hydrolysis;
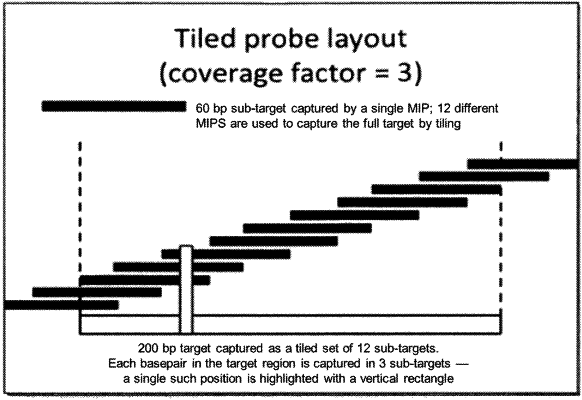
introducing a plurality of molecular inversion probes (MIPs) to the plurality of fragments, wherein each of the plurality of MIPs hybridize to the fragments and captures a different one of a plurality of overlapping subregions of the same strand of a fragment, wherein the fragmenting step promotes hybridization of the MIPs to the fragments;
circularizing the hybridized MIPs;
amplifying the circularized MIPs to generate amplicons; and
sequencing the amplicons.
|