| CPC G16B 50/50 (2019.02) [G06F 16/1744 (2019.01); G16B 30/00 (2019.02); G16B 30/10 (2019.02); G16B 40/00 (2019.02); G16B 50/00 (2019.02); H03M 7/3059 (2013.01)] | 16 Claims |
|
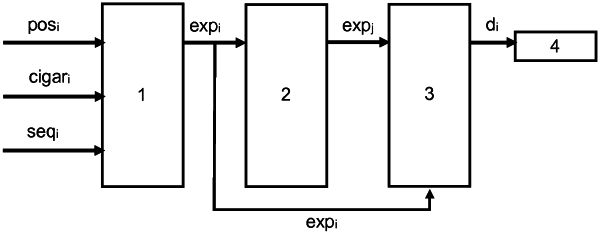
1. A computer implemented method for reducing computer storage size requirements for and transmission costs for transmitting genomic data, via a compressing of genomic data stored in at least one data file as a plurality of reads built by a genome sequencing method, each read including a read mapping position, a read Compact Idiosyncratic Gapped Alignment Report (CIGAR) string, comprising a compressed pairwise alignment format report defining a sequence of matches/mismatches and deletions or gaps, and a read actual sequenced nucleotide sequence that is a local part of a donor genome, the computer implemented method comprising:
a sequence of computer processing steps, including:
a computer processing step of initializing a window buffer memory,
a computer processing step of selecting one of the reads as a current read,
a computer processing step of expanding the current read to an unwound nucleotide sequence of the current read, using the mapping position and the CIGAR string of said current read,
a computer processing step, conditional on no current storage in the window buffer memory of any previous read-based unwound nucleotide sequences, comprising
computing an initial read difference, passing said initial read difference to a computer-implemented entropy encoder,
a computer processing step, by the computer implemented entropy encoder, upon receiving said computed initial read difference, of compressing said computer initial read difference to a compressed initial read difference, and replacing encoding said current read by the compressed initial difference, wherein the initial read difference is based on the unwound nucleotide sequence of the current read and another data,
a computer processing step, conditional on a current storage in the window buffer memory of at least one previous read-based unwound nucleotide sequences, each corresponding to a respective expanding of a different previous read, comprising:
computing a computed difference between the unwound nucleotide sequence of said current read and each of said at least one previous read-based unwound nucleotide sequences,
selecting, as a selected difference, a one from said at least one computer differences that indicates a minimum,
passing said computed difference to a computer-implemented entropy encoder, and including in said computed difference, the difference of the current read mapping position and at least one previous read mapping positions and the difference of the current read unwound nucleotide sequence and at least one previous read unwound nucleotide sequences, and
a computer processing step, by the computer implemented entropy encoder, upon receiving said computed difference, of entropy encoding compressing said computed difference to a compressed difference, and replacing said current read by the compressed difference, and
repeating said sequence of computer steps, using for each repeat of the sequence the current read as one of said previous reads and a following read as a new current read, until no more following reads are available,
wherein, in the repeating said sequence of computer processing steps, upon said current read having a predetermined number of consecutive previous reads, the predetermined number being at least two:
the step of computing the computed difference includes computing at least two computed differences, each a respective computed difference between the unwound nucleotide sequence of said current read and an unwound nucleotide sequence of a corresponding one among each of at least two of said consecutive reads and selecting, as a selected difference, a one from said at least two computer differences that indicates a minimum, and
the computer processing step of passing said computed difference to said entropy encoder is configured to pass said selected difference.
|