| CPC C12Q 1/6869 (2013.01) [C12N 15/1093 (2013.01); C12Q 1/6883 (2013.01); C12Q 1/6886 (2013.01); C12Q 2600/118 (2013.01); C12Q 2600/156 (2013.01)] | 19 Claims |
|
1. A method of detecting genomic location of a transposon in a subject, comprising
a) providing isolated DNA sample from a test sample of the subject;
b) fragmenting the DNA sample to obtain DNA fragments;
c) ligating adaptors to both ends of the DNA fragments to obtain a transposon-adaptor library;
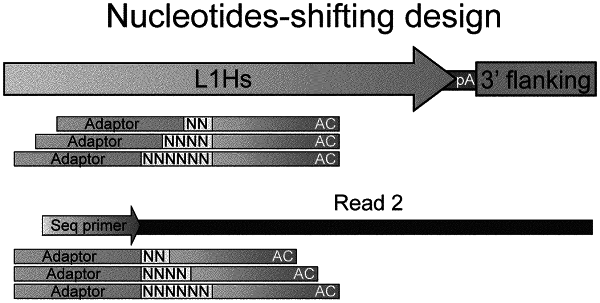
d) amplifying the transposon-adaptor library with primers comprising a first primer and a second primer, wherein the first primer comprises a transposon specific sequence capable of hybridizing to an internal sequence of the transposon, a second primer comprises an adapting sequence capable of hybridizing to the adaptor, thereby amplifying the transposon and flanking sequences thereof;
e) sequencing the obtained product from amplification to obtain the flanking sequences; and
f) aligning the obtained flanking sequences with a reference genomic sequence to obtain genomic location and orientation of the transposon; and
wherein the first primer is a primer set comprising 3 or more different first primers, wherein each of the first primer further comprises a library adapting sequence at its 5′ end and a spacer region is provided between the library adapting sequence and the transposon sequence, and wherein the length of the spacer region in each first primer is different.
|