| CPC G06T 11/206 (2013.01) [G06F 3/0481 (2013.01); G06F 3/04842 (2013.01); G06F 3/14 (2013.01); G06F 16/245 (2019.01); G06N 5/04 (2013.01); G06N 7/01 (2023.01); G06N 20/00 (2019.01); G06T 11/001 (2013.01); G06T 11/203 (2013.01); G06T 2200/24 (2013.01)] | 17 Claims |
|
1. A method, implemented using a computer system that includes one or more processors and system memory, the method comprising:
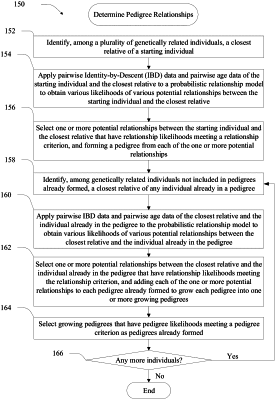
iteratively growing, by the one or more processors, pedigrees for a plurality of genetically related individuals, wherein the pedigrees are grown based on pairwise identity-by-descent (IBD) data between the genetically related individuals, and wherein the pedigrees are selected for growth based on a configurable pedigree likelihood;
combining, by the one or more processors, pairs of pedigrees that share a greatest amount of common IBD data above a pre-determined threshold;
storing, by the one or more processors and in a database, pedigree relationship data of the plurality of genetically related individuals, wherein the pedigree relationship data is based on the pedigrees grown and combined and comprise a plurality of entries representing the plurality of genetically related individuals, wherein child entries of the plurality of entries comprise links relating to parent entries of the plurality of entries, wherein each child entry and its parent entries respectively represent a child and its biological parents;
determining, by the one or more processors, a set of root entries without known parents in the plurality of entries and from which all of the plurality of entries are reachable via the links;
for each respective root entry of the set of root entries, forming, by the one or more processors, a respective subtree of a plurality of subtrees by positioning nodes in each of the respective subtrees, wherein the positioning comprises:
using the respective root entry as a root node,
identifying entries reachable from the root entry by traversing the links from parent entries to child entries and traversing between partner entries, wherein partner entries are two parent entries of the same child entry, and
positioning the entries that are reachable as nodes in the respective subtree, wherein each node in the respective subtree represents an entry related to the root node;
merging, by the one or more processors, the plurality of subtrees to form a pedigree graph for the plurality of genetically related individuals, comprising:
identifying corresponding nodes on different subtrees within the plurality of subtrees, wherein corresponding nodes are nodes that represent common individuals, and
combining the corresponding nodes such that the subtrees containing the corresponding nodes become one tree; and
displaying, by the one or more processors, the pedigree graph.
|