| CPC C12Q 1/6881 (2013.01) [C07K 7/04 (2013.01); G01N 33/574 (2013.01); G01N 33/6878 (2013.01); G16B 5/00 (2019.02); G16B 5/20 (2019.02); G16B 15/00 (2019.02); G16B 15/30 (2019.02); G16B 20/00 (2019.02); G16B 20/20 (2019.02); G16B 20/30 (2019.02); G16B 25/10 (2019.02); G16B 30/00 (2019.02); G16B 30/10 (2019.02); G16B 40/00 (2019.02); G16B 45/00 (2019.02); G16B 50/10 (2019.02); G16B 50/20 (2019.02); C12Q 2600/156 (2013.01)] | 8 Claims |
|
1. A method of producing a peptide for treating, monitoring, or diagnosing a disease in a subject, the method comprising the steps of:
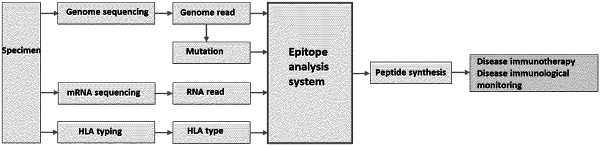
A) inputting into an analyzer information related to a mutation specific to a diseased tissue of the subject and information on an MHC type of the subject, wherein the step A) comprises the steps of:
A-1) making the analyzer sequence a genome of the subject to obtain and map the information related to a genome read of the subject and the mutation thereof, and then search for the mutation specific to the diseased tissue to obtain the mutation specific to the diseased tissue; and
A-2) making the analyzer sequence an RNA of the subject to obtain information on an RNA read of the subject, map an RNA read of the diseased tissue, and search for a mutation, and/or derive an amount of expression, and map an RNA read of a normal tissue to search for a somatic cell mutation and/or derive an amount of expression to compare said amount with the amount of expression derived based on the RNA read of the diseased tissue;
B) making the analyzer analyze an epitope associated with the mutation based on the information related to the mutation specific to the diseased tissue, the information on the MHC type, and information on the disease,
wherein the step B) comprises the step of making the analyzer add an annotation for the mutation specific to the diseased tissue based on a reference information database to identify a candidate mutation, wherein nucleic acid information of the candidate mutation is then converted to amino acid information to produce a wild-type (WT) peptide and a mutant (MT) peptide, and then the analyzer is made to search for an epitope using the MHC type, the WT peptide, and the MT peptide after which epitopes are ranked, and to output an epitope list;
wherein the ranking is performed by taking into consideration at least one element selected from the group consisting of prioritization of the mutation, presence/absence of gene expression, and prioritization of a peptide; and
wherein the ranking is sorted by applying, in order, a value of IC50 between HLA-peptide, the number of epitope searching programs which have found a hit, and the number of mutation searching softwares which have found a hit; and
C) producing the peptide based on at least the epitope list output from the analyzer in accordance with the ranking of step B).
|