| CPC G16B 30/10 (2019.02) [G06F 16/285 (2019.01); G16B 40/00 (2019.02); G16B 30/20 (2019.02)] | 16 Claims |
|
1. A system for improving accuracy of polynucleotide sequencing, the system comprising:
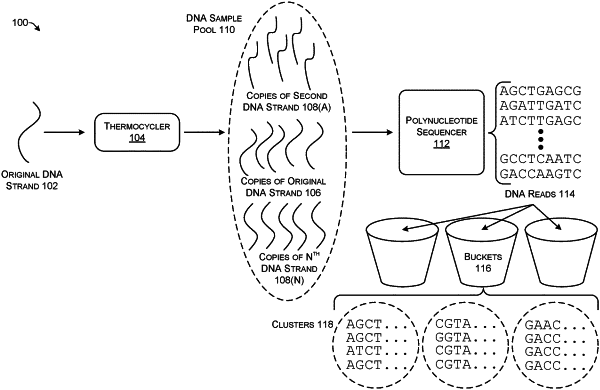
a polynucleotide sequencer configured to generate a plurality of DNA reads from a plurality of DNA strands having different nucleotide sequences, wherein the plurality of DNA reads includes errors introduced by the polynucleotide sequencer;
at least one processing unit;
a memory in communication with the processing unit;
a clusterization module stored in the memory and executable on the processing unit to divide the plurality of DNA reads into clusters by first grouping DNA reads having a same hash as determined by randomized locality-sensitive hashing (LSH) into buckets and then grouping DNA reads in a same bucket into clusters based at least in part on similarity of signatures of the DNA reads, wherein the signatures are generated by a technique that deterministically embeds edit-distance space into Hamming space; and
a consensus output sequence generator stored in the memory and executable on the processing unit configured to generate a single consensus output sequence from each cluster that has less error than individual reads in each cluster.
|