| CPC C12Q 1/6869 (2013.01) [G16B 30/10 (2019.02); G16B 30/20 (2019.02); G16B 50/30 (2019.02)] | 20 Claims |
|
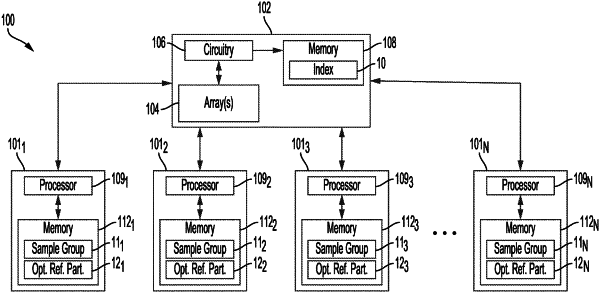
1. A method of processing a plurality of sample reads for genome sequencing using a system comprising at least one systolic array including a plurality of groups of cells, the method comprising:
for each sample read of the plurality of sample reads:
comparing substring sequences from the sample read to reference sequences representing different portions of a reference genome, the comparing comprising:
for each substring sequence from the sample read:
storing the substring sequence in a first group of cells of the plurality of groups of cells of the at least one systolic array, wherein the first group of cells stores the substring sequence and further stores a first reference sequence;
comparing the substring sequence stored in the first group of cells to the first reference sequence stored in the first group of cells; and
passing sample values forming the substring sequence from the first group of cells to a second group of cells of the plurality of groups of cells of the at least one systolic array for comparison of the substring sequence to a second reference sequence stored in the second group of cells, wherein the second reference sequence is different from the first reference sequence and the second group of cells does not include any cells of the first group of cells;
identifying one or more reference sequences stored in one or more groups of cells of the plurality of groups of cells of the at least one systolic array that match one or more substring sequences compared to the one or more reference sequences; and
determining a probabilistic location of the sample read within the reference genome based on the one or more identified reference sequences that match the one or more compared substring sequences; and
sorting the plurality of sample reads into a plurality of sample groups based at least in part on the determined probabilistic locations of the respective sample reads.
|