| CPC G06T 7/0012 (2013.01) [G06T 7/10 (2017.01); G06T 2207/20081 (2013.01); G06T 2207/20132 (2013.01); G06T 2207/30068 (2013.01)] | 2 Claims |
|
1. A system for automatically identifying mitosis in hematoxylin-eosin (H&E) stained breast cancer pathological images, comprising:
an input image preprocessing module: cutting an original picture into a predetermined patch size, and performing a data enhancement through picture flipping and rotation;
the original picture obtains a mask binary image through a pixel position labeling;
obtaining training data of a segmentation network using a data set labeled at a pixel level, obtaining a corresponding mask binary image through the pixel position labeling, and cutting a corresponding RGB image based on the mask binary image to obtain mitotic patches and non-mitotic patches as the training data;
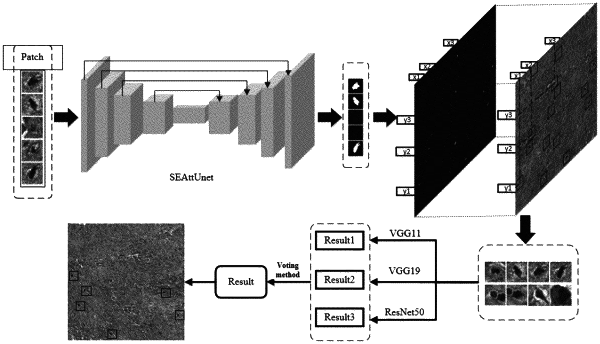
a segmentation module: training the segmentation network by cutting mitotic patches and non-mitotic patches from a training set, cutting data of a test set according to a corresponding size and sending it to the segmentation network to obtain a patch-level segmentation result, and then reconstructing a segmented result into an image of the original size according to patch coordinate information intercepted in a preprocessing stage of it the segmentation module;
a classification module: training three classification networks, performing a decision-making level fusion on results of the three classification networks, using a voting method to obtain a final output result, and then restoring and marking the final output result to obtain a final detection result;
the segmentation module comprises an attention segmentation network module and a candidate cell selection method module;
the classification module comprises a three-branch classification network module, a decision-making fusion algorithm module and a classification result marking module; and
the attention segmentation network module introduces an SE (Squeeze-and-Excitation) module:
constructing a segmentation network based on Attention U-Net, combining an attention mechanism to realize a weighted extraction of different features in spaces and channels, aiming at morphological features of research objectives mitotic cells, improving a squeeze operation in the SE module, and taking into account both texture information and background information; and based on above characteristics, enabling the segmentation network SEAttUnet based on the Attention U-Net to realize a redistribution of attentions of space positions in different channels and feature maps.
|