| CPC G16B 40/20 (2019.02) [G16B 20/10 (2019.02); G16B 20/20 (2019.02); G16B 25/00 (2019.02); G16B 40/00 (2019.02)] | 20 Claims |
|
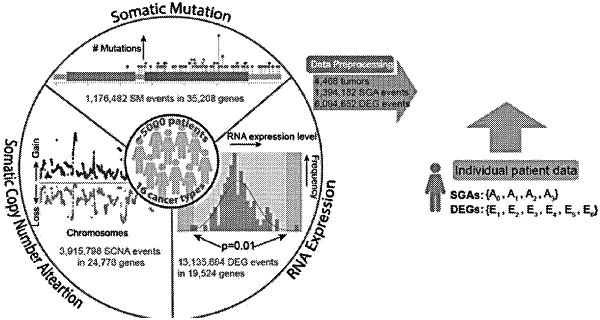
1. A method for detecting a somatic genome alteration (SGA) with tumor-specific functional impact in a genome of a specific tumor t of a subject, comprising:
receiving a training dataset that comprises gene expression data indicating respective levels of expression of a plurality of genes in a collection of tissue samples including tumors;
receiving a set of SGAs in the genome of t;
training a bipartite causal Bayesian network (CBN) with maximal posterior probability, including:
identifying a set of differentially expressed genes (DEGs) in the genome of t by designating each gene in a set of genes in the genome of t as a DEG if the respective level of expression of the gene in t falls outside a significance boundary determined based on respective levels of expression of the gene in the collection of tissue samples represented in the training dataset;
for each DEG in the set of DEGs:
(i) identifying, from among multiple SGAs in the set of SGAs that are possible causes of the DEG, one SGA that is the most probable cause of the DEG, and
(ii) providing the DEG with just a single causal edge in the CBN with maximal posterior probability, the single causal edge connecting the DEG in the CBN to the SGA that has been identified as the most probable cause of the DEG, wherein the DEG is not connected in the CBN to any other SGAs except for the SGA that has been identified as the most probable cause of the DEG; and
identifying a particular SGA that is connected by respective causal edges to at least a threshold number of DEGs in the CBN with maximal posterior probability as the SGA with tumor-specific functional impact in the genome of t.
|