| CPC G06T 7/0012 (2013.01) [G06T 2207/10024 (2013.01); G06T 2207/10068 (2013.01); G06T 2207/20081 (2013.01); G06T 2207/30032 (2013.01)] | 3 Claims |
|
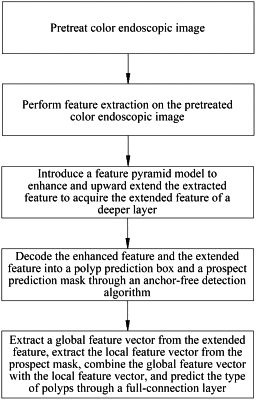
1. A method for detection and pathological classification of polyps via colonoscopy based on an anchor-free technique, comprising the following steps:
pretreating a color endoscopic image;
performing feature extraction on the color endoscopic image that is pretreated;
introducing a feature pyramid model to enhance the performing of feature extraction on the color endoscopic image that is pretreated to acquire an enhanced feature, and upward extending the enhanced feature to acquire an extended feature of a deeper layer;
decoding feature information of the enhanced feature and the extended feature through an anchor-free detection algorithm to acquire a polyp prediction box and a prospect prediction mask;
extracting a global feature vector from the extended feature and extracting a local feature vector from the prospect prediction mask, and combining the global feature vector with the local feature vector, to predict a type of polyps through a full-connection layer;
wherein the steps of performing feature extraction on the color endoscopic image that is pretreated are as follows:
using ResNeXt101 pre-trained based on an ImageNet as a backbone network;
dividing the ResNeXt101 into 5 different stages of R1, R2, R3, R4, and R5 with a maxpool as a boundary;
with a deepening of the backbone network, reducing the size of a feature map acquired after each pooling by a half, and doubling the number of channels; and
extracting the network outputs of C2 to C5 in the 4 different stages of R2 to R5 as the extracted feature map;
the steps of introducing a feature pyramid model to enhance the feature extracted to acquire an enhanced feature, and upward extending the enhanced feature to acquire an extended feature of a deeper layer are as follows:
enhancing semantic information of a shallow feature layer by using a top-down method of a feature pyramid structure to acquire shallow feature maps P2, P3, P4, and P5 having deep information, and meanwhile upward extending the feature pyramid structure by one layer to acquire a semantic information feature map P6 of the deeper layer;
the steps of decoding the feature information of the enhanced feature and the extended feature through an anchor-free detection algorithm to acquire a polyp prediction box and a prospect prediction mask are as follows:
S100: for feature points of different feature layers in a training stage, giving different labels according to the sizes of target polyps and allocating the labels to different scales of feature layers as actual labels, wherein object box information with actual label is used for a regression function to calculate positions of a candidate boxes;
S101: acquiring H×W×4 tensor for position prediction, a H×W×1 dimensional prospect prediction mask and a H×W×1 tensor Center-ness for measuring the degree to which a current pixel is offset from the center point of a real target;
S102: performing position information decoding on the H×W×4 output tensor,
wherein the output of a feature point x, y on the feature map is predicted as [l*,r*,t*,b*], l*,r*,t* and b* are respectively distances from the feature point x, y to left, right, upper and lower sides of the polyp prediction box,
x0=x−l*,
y0=y−t*,
w=l*−r*, and
h=t*+b*,
the position of the predicted polyp is [x0,y0,w,h] via decoding, wherein x0, y0 are coordinates of an upper left corner of the polyp prediction box, and w and h are respectively a width and height of the polyp prediction box;
S103: performing distance thermal value calculation on position information regression value corresponding to each feature point on a H×W×1 tensor output by Center-ness to help a target judgment of the feature point on the polyp at the current position, wherein the specific formula is as follows:
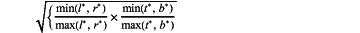 wherein, min (x,y) is a minimal value of x and y, and similarly max (x, y) is a maximum value of x, y,
in the stage of training, calculating a loss value by using a distance thermogram and the H×W×1 tensor output by Center-ness, and utilizing a two-class cross-entropy function as a loss function,
when the feature point is closer to the center of the prediction box, the value of the loss function is smaller, conversely, the value of the loss function is larger;
the steps of extracting the global feature vector from the extended feature are as follows:
performing an average pooling operation on the extended feature of the feature pyramid to acquire a 256×1 dimensional global feature vector, wherein the extended feature degree has a stride of 128;
the steps of extracting the local feature vector from the prospect prediction mask are as follows:
S200: introducing a prospect attention mechanism, superimposing the outputs of the prospect prediction mask and the corresponding feature map of the feature pyramid that are convoluted, and then retaining a prospect part of the feature map corresponding to the prospect mask, whereas ignoring a background part, to acquire the local feature map; wherein the calculation formula is as follows:
Mlocal=M*a
M is the feature map output by the feature pyramid, a is the prospect mask, * is an array element product, and Mlocal is the local feature map;
S201: applying a global average pooling operation on all local feature maps to acquire a 256×1 dimensional local feature vector; and
the steps of combining the local feature vector with the global feature vector to predict the type of polyps through a full-connection layer are as follows:
combining 5 256×1 dimensional local feature vectors to generate a 1280×1 local feature vector, then performing dimensionality reduction on the 1280×1 local feature vector via a 1×1×256 convolution layer to acquire a 256 dimensional local feature vector, then combining the local feature vector with the global feature vector as a 512×1 dimensional feature vector, and finally predicting the types of polyps through a full-connection layer.
|