| CPC G16H 50/20 (2018.01) [G16B 25/10 (2019.02); G16H 10/40 (2018.01); G16H 10/60 (2018.01); G16H 50/30 (2018.01); G16H 50/50 (2018.01); G16H 50/70 (2018.01); G16H 70/60 (2018.01)] | 6 Claims |
|
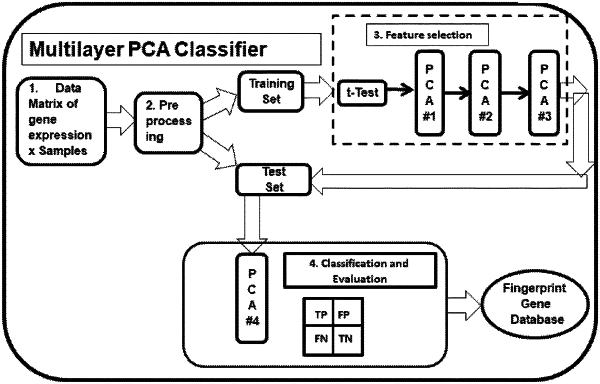
1. A method of diagnosing disease in a patient's biological sample using a computer-implemented multilayer Principal Component Analysis (PCA) Classifier comprising:
obtaining gene expression profile data, comprising gene expression values of human biological samples with known disease/disease stages and control/healthy characteristics, also known as true positives/controls patient biological samples;
preprocessing the gene expression profile data;
generating using the preprocessed gene expression profile data, training samples comprising a fraction of the true positives/controls patient biological samples, and test/validation samples comprising another fraction of the true positives/controls patient biological samples;
generating using the preprocessed gene expression profile data, a training set comprising a matrix of genes by training samples;
performing a t-test on the training set, and generating a new training set comprising a matrix of genes by training samples as input to a first PCA layer;
selecting, using the first PCA layer, genes to add to a Gene Pool A by performing a linear transformation on the new training set to generate a matrix of gene scores, wherein genes with an absolute value gene score above a variable threshold between 60% to 85% of a maximum gene score in the matrix of gene scores are added to Gene Pool A;
generating, using a transpose of the new training set, a matrix of training samples by genes, as input to a second PCA layer;
selecting, using the second PCA layer, genes to add to a Gene Pool B by performing a linear transformation on the transpose of the new training set, to generate first principal component vectors comprising coefficients of linear combinations of the genes, wherein genes with absolute value coefficients greater than 1/(sqrt(total number of gene coefficients in leading principal component vectors)) are added to Gene Pool B;
identifying disease specific fingerprint genes by determining genes common to Gene Pool A and Gene Pool B;
generating, using the disease specific fingerprint genes, a matrix of disease specific fingerprint genes by training samples;
classifying, using a third PCA layer, expression levels of the disease specific fingerprint genes by performing a linear transformation on the matrix of fingerprint genes by training samples, to generate a matrix of gene scores, to identify fingerprint genes with positive valued gene scores as upregulated, and to identify fingerprint genes with negative valued gene scores as downregulated;
identifying, using the expression values of the upregulated or downregulated disease specific fingerprint genes, treatment plans and medications to treat the specific disease;
generating, using the disease specific fingerprint genes and the test/validation samples, a matrix of disease specific fingerprint genes by test/validation samples;
identifying, using a fourth PCA layer, the test/validation samples as disease or healthy, by performing linear transformation on a matrix of test/validation samples by disease specific fingerprint genes, to create principal component vectors, wherein two largest variance principal component vectors are selected and multiplied with the matrix of the disease specific fingerprint genes by test/validation samples, to create a graphical plot on a computer display screen to distinguish the disease test/validation samples from healthy test/validation samples, wherein the test/validation samples are classified as disease/healthy with a certain set accuracy;
generating, using the four PCA layers, disease specific fingerprint genes for a plurality of diseases;
generating using the four PCA layers, a plurality of matrices comprising gene expression profile data of disease specific fingerprint genes by disease specific true positive patient biological samples for the plurality of diseases;
generating a fingerprint gene data matrix comprising the plurality of matrices of gene expression profile data of disease specific fingerprint genes by disease specific true positive patient biological samples, and a fingerprint gene library comprising the disease specific fingerprint genes for the plurality of diseases;
generating a fingerprint gene database comprising the fingerprint gene data matrix and the fingerprint gene library for identifying disease in patients, and administering medications;
obtaining a test patient's biological sample from comprising gene expression profile data of the test patient's biological sample, using an invasive procedure;
generating using each matrix of gene expression profile data of disease specific fingerprint genes by disease specific true positive patient biological samples from the fingerprint gene data matrix and the test patient's gene expression profile data, a plurality of matrices to screen the test patient's biological sample for disease using the plurality of matrices thus generated, wherein each matrix in the plurality of matrices screens the test patient's biological sample for a specific disease;
identifying, using the fourth PCA layer, presence of a specific disease in the test patient's biological sample, by performing linear transformation on each matrix in the plurality of matrices of gene expression profile data of disease specific fingerprint genes by disease specific true positive patient biological samples, to create principal component vectors, wherein two largest variance principal component vectors are selected to create graphical plots on a computer display screen that classifies the test patient's biological sample as disease or healthy;
administering, using the expression values of the upregulated or downregulated disease specific fingerprint genes, treatment plans and medications to treat the specific disease in the test patient's biological sample.
|