| CPC G16B 10/00 (2019.02) [G06F 16/22 (2019.01); G06F 16/24 (2019.01); G16B 20/20 (2019.02); G16B 30/00 (2019.02)] | 19 Claims |
|
1. A genealogical indexing system comprising:
a processor;
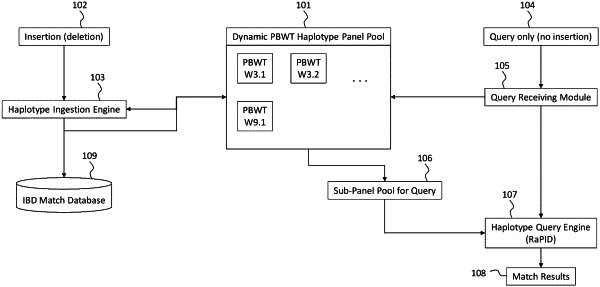
a haplotype panel pool in electronic communication with a non-transitory computer-readable medium that is operably coupled to the processor, the haplotype panel pool including a plurality of panels that is indexed by a positional Burrows-Wheeler transform (PBWT) representation, each of the plurality of panels including a plurality of haplotypes, the haplotype panel pool having a set of variant sites, wherein each of the plurality of haplotypes is indexed by the PBWT representation in a reversed prefix order; and
the non-transitory computer-readable medium having computer-readable instructions stored thereon that, when executed by the processor, cause the genealogical indexing system to automatically identify a genealogical match with an input genetic sequence by executing instructions comprising:
selecting, via a computing nodule in electronic communication with the haplotype panel pool, a minimum identity-by-descent segment length value of at least one million base pairs, such that haplotype matches greater than or equal to the minimum identity-by-descent segment length indicate a match between the input genetic sequence and the haplotype panel pool, and such that haplotype matches lesser than the minimum identity-by-descent segment length indicate a lack of a match between the input genetic sequence and the haplotype panel pool;
selecting, via the computing nodule in electronic communication with the haplotype panel pool, a plurality of portions of each of the plurality of panels to sample each of the plurality of panels;
projecting, via the computing nodule in electronic communication with the haplotype panel pool, the sample to the set of variant sites for each of the plurality of panels in the haplotype panel pool, such that the sample can be compared against the samples in the haplotype panel pool via the set of variants;
receiving, via the computing nodule in electronic communication with the haplotype panel pool, the input genetic sequence;
automatically indexing, via a haplotype querying engine of the computing nodule in electronic communication with the haplotype panel pool, executable by the processor, the input genetic sequence by the PBWT representation, thereby transforming the input genetic sequence into a PBWT indexed input genetic sequence including the reversed prefix order;
activating, via the haplotype querying engine of the computing nodule in electronic communication with the haplotype panel pool, an amount of the plurality of panels from the haplotype panel pool for testing;
comparing, via the haplotype querying engine of the computing nodule in electronic communication with the haplotype panel pool, the PBWT indexed input genetic sequence with the amount of the plurality of panels by comparing the PBWT indexed input genetic sequence with the set of variant sites; and
via the haplotype querying engine of the computing nodule in electronic communication with the haplotype panel pool, automatically identifying, in real-time and independent of a total number of haplotypes in the plurality of haplotypes, the genealogical match between the input genetic sequence and the haplotype pool by:
based on a determination that the PBWT indexed input genetic sequence matches the amount of the plurality of panels by a value greater than or equal to the minimum identity-by-descent segment length of at least one million base pairs, determining the genealogical match between the input genetic sequence and the haplotype panel pool; and
based on a determination that the PBWT indexed input genetic sequence does not match the amount of the plurality of panels by a value greater than or equal to the minimum identity-by-descent segment length of at least one million base pairs, determining a lack of the genealogical match between the input genetic sequence and the haplotype panel pool.
|