| CPC G16B 20/20 (2019.02) [C12Q 1/6827 (2013.01); C12Q 1/6869 (2013.01); G16B 20/10 (2019.02); G16B 30/00 (2019.02); G16B 30/10 (2019.02); G16B 30/20 (2019.02); G16B 50/20 (2019.02); G16B 50/30 (2019.02); G16B 20/00 (2019.02)] | 21 Claims |
|
1. A sequence analysis method for analyzing a nucleic acid sequence, the sequence analysis method comprising:
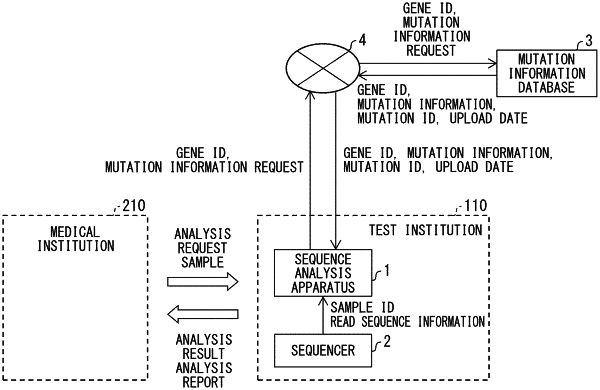
obtaining, by a processing device from a sequencer device, a first plurality of read sequences which are read from a first nucleic acid sequence, wherein the first plurality of read sequences comprise 40 to 200 giga-bases (Gb); and
at a first time, determining, by the processing device, the first nucleic acid sequence by aligning the first plurality of read sequences with reference to a single reference sequence, wherein
the single reference sequence comprises at least a first rearrangement sequence and a second rearrangement sequence that is different from the first rearrangement sequence, wherein an entirety of the second rearrangement sequence is appended after an entirety of the first rearrangement sequence within the single reference sequence to form a continuous string including at least the first rearrangement sequence and the second rearrangement sequence;
the method further comprising:
at a second time after the first time, determining that a new known mutation information exists in a mutation information database, wherein a creation date for the new known mutation information corresponds to a time that is after a creation date for the single reference sequence;
updating the reference sequence to add a third rearrangement sequence related to the new known mutation information by appending an entirety of the third rearrangement sequence after at least one of the entirety of the first rearrangement sequence or the entirety of the second rearrangement sequence within the single reference sequence to create an updated single reference sequence that is formed as a continuous string including at least the first rearrangement sequence, the second rearrangement sequence, and the third rearrangement sequence;
obtaining, by the processing device from the sequencer device, a second plurality of read sequences which are read from a second nucleic acid sequence; and
determining, by the processing device, the second nucleic acid sequence by aligning the second plurality of read sequences with reference to the updated single reference sequence.
|