| CPC G16H 20/10 (2018.01) [G16B 5/00 (2019.02)] | 24 Claims |
|
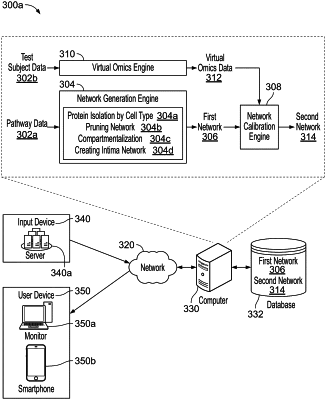
23. A method of screening a potential subject for enrollment in a clinical trial testing safety or efficacy, or both, of a candidate anti-diabetic agent for atherosclerotic cardiovascular disease, the method comprising:
receiving a first input of non-invasively obtained imaging data related to a plaque from a set of test subjects;
receiving a second input of molecular expression data from the set of test subjects;
creating a training set comprising the first input and the second input;
training a neural network using the training set, the neural network configured to predict molecule levels based on the non-invasively obtained imaging data from the set of test subjects;
receiving non-invasively obtained imaging data related to a plaque from the potential subject;
generating virtual ′omics data that include predicted molecule levels of the potential subject, by applying the neural network to the non-invasively obtained imaging data from the potential subject;
providing the virtual ′omics data to a systems biology model of atherosclerotic cardiovascular disease to generate a subject-specific systems biology model, wherein
(i) the systems biology model represents a plurality of pathways associated with atherosclerotic cardiovascular disease,
(ii) each pathway in the plurality of pathways corresponds to one or more of MTOR, NFκβ1, ICAM1, or VCAM1,
(iii) the systems biology model includes a disease-associated molecule level for each molecule in the systems biology model, and
(iv) the subject-specific systems biology model includes predicted molecule levels that are updated from the disease-associated molecule level;
updating the subject-specific systems biology model with predicted molecular levels derived from information relating to an effect on glucose by a candidate anti-diabetic agent based on a known mechanism of action of the candidate anti-diabetic agent; and
simulating a therapeutic response by the potential subject to the candidate anti-diabetic agent in the updated subject-specific systems biology model to obtain a simulated therapeutic effect, wherein the simulated therapeutic effect is based on change in the predicted molecule levels in the updated subject-specific systems biology model with and without the candidate anti-diabetic agent.
|