| CPC G16H 50/20 (2018.01) [G16H 50/30 (2018.01)] | 7 Claims |
|
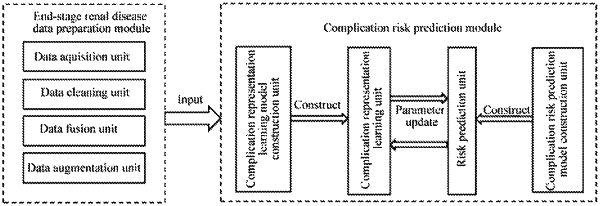
1. A system for predicting end-stage renal disease complication risk based on contrastive learning, comprising:
an end-stage renal disease data preparation module, configured to extract structured data of a patient using a hospital electronic information system and daily monitoring equipment, and process the structured data to obtain augmented structured data;
the end-stage renal disease data preparation module comprising:
a data augmentation unit, configured to obtain the augmented structured data using a data augmentation method combining propensity score matching with Synthetic Minority Oversampling Technique (SMOTE) for original fusion features obtained by processing the structured data;
the data augmentation unit comprising:
a fusion feature component, configured to take a patient with an end-stage renal disease complication as a positive sample, take a patient with no the end-stage renal disease complication as a negative sample, represent the positive sample and the negative sample with the original fusion features, and perform normalization operation on the original fusion features of the positive sample and the negative sample, to obtain a fusion feature;
a propensity score component, configured to select one dimension of the fusion feature arbitrarily to serve as an intervening variable, with other dimensions of the fusion feature serving as a concomitant variable set, to obtain a propensity score through loss function optimization;
a matching component, configured to make all positive samples constitute a positive sample universal set, make all negative samples constitute a negative sample universal set, and make the positive sample universal set match negative sample subsets in the negative sample universal set based on the propensity score;
a positive sample augmentation component, configured to obtain an augmented positive sample by performing a SMOTE algorithm on the positive sample universal set, wherein the positive sample universal set and the augmented positive sample constitutes a positive sample augmented set;
a negative sample augmentation component, configured to obtain an augmented negative sample by performing a SMOTE algorithm on the negative sample subsets, wherein the negative sample subsets and the augmented negative sample constitutes a negative sample augmented set; and
an augmentation component, configured to make the positive sample augmented set and the negative sample augmented set jointly constitute the augmented structured data; and
a complication risk prediction module, configured to construct a complication representation learning model and a complication risk prediction model, and perform training and learning on the augmented structured data through the complication representation learning model to obtain a complication representation, and perform end-stage renal disease complication risk prediction using the complication representation through the complication risk prediction model;
wherein parameter construction of a total loss function of the complication representation learning model comprises a normalization representation, and a determining mode of the normalization representation comprises: inputting the augmented structured data in pairs into an encoder of the complication representation learning model, to obtain an initial complication representation, obtaining a contrastive representation from the initial complication representation through a projector of the complication representation learning model, and obtaining the normalization representation from the contrastive representation through feature normalization operation.
|