| CPC C12Q 1/6886 (2013.01) [C12N 15/1065 (2013.01); C12N 15/1096 (2013.01); C12Q 1/6806 (2013.01); C12Q 1/6869 (2013.01); C12Q 2600/158 (2013.01); C12Q 2600/16 (2013.01)] | 23 Claims |
|
1. A method of detecting genomic alteration and/or detecting gene expression and/or quantifying a level of gene expression using RNA in a biological sample, comprising:
(a) extracting RNA from the biological sample and converting the RNA to complementary DNA (cDNA);
(b) performing a plurality of multiplexed PCR reactions on the converted cDNA using
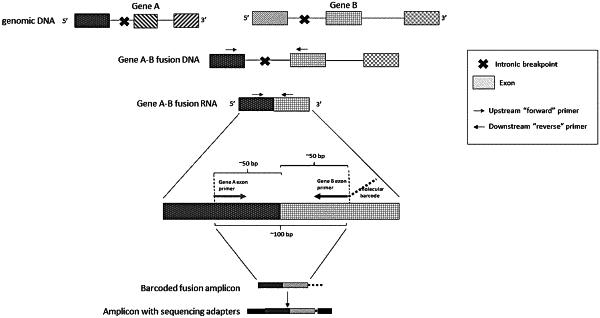
(I) a plurality of forward and reverse primer pairs specific to a plurality of target genes that are capable of undergoing genomic alteration,
wherein each forward primer of the plurality of forward and reverse primer pairs specific to the plurality of target genes that are capable of undergoing genomic alteration is complementary to a sequence located about 50 base pairs upstream of an exonic junction of each target gene that is capable of undergoing genomic alteration,
wherein each reverse primer of the plurality of forward and reverse primer pairs specific to the plurality of target genes that are capable of undergoing genomic alteration is complementary to a sequence located about 50 base pairs downstream of an exonic junction of each target gene that is capable of undergoing genomic alteration,
wherein each reverse primer of the plurality of forward and reverse primer pairs specific to the plurality of target genes that are capable of undergoing genomic alteration comprises a barcode sequence on its 5′ end, wherein the barcode sequence of each reverse primer corresponding to each target gene that is capable of undergoing genomic alteration is different, and/or
(II) a plurality of forward and reverse primer pairs specific to a plurality of control housekeeping genes, wherein:
(i) each forward primer of the plurality of forward and reverse primer pairs specific to the plurality of control housekeeping genes is complementary to a sequence spanning an exon-exon junction of each control housekeeping gene,
wherein each reverse primer of the plurality of forward and reverse primer pairs specific to the plurality of control housekeeping genes is complementary to a sequence about 100 base pairs downstream of the sequence spanning the exon-exon junction of each control housekeeping gene,
wherein each reverse primer of the plurality of forward and reverse primer pairs specific to the plurality of control housekeeping genes comprises a barcode sequence on its 5′ end, wherein the barcode sequence of each reverse primer corresponding to each control housekeeping gene is different;
(ii) each reverse primer of the plurality of forward and reverse primer pairs specific to the plurality of control housekeeping genes is complementary to a sequence spanning an exon-exon junction of each control housekeeping gene,
wherein each forward primer of the plurality of forward and reverse primer pairs specific to the plurality of control housekeeping genes is complementary to a sequence about 100 base pairs downstream of the sequence spanning the exon-exon junction of each control housekeeping gene,
wherein each reverse primer of the plurality of forward and reverse primer pairs specific to the plurality of control housekeeping genes comprises a barcode sequence on its 5′ end, wherein the barcode sequence of each reverse primer corresponding to each control housekeeping gene is different; or
(iii) each forward and each reverse primer of the plurality of forward and reverse primer pairs specific to the plurality of control housekeeping genes is complementary to consecutive sequences spanning an exon-exon junction of each control housekeeping gene,
wherein each reverse primer of the plurality of forward and reverse primer pairs specific to the plurality of control housekeeping genes comprises a barcode sequence on its 5′ end, wherein the barcode sequence of each reverse primer corresponding to each control housekeeping gene is different, and/or
(III) a plurality of primer sets specific to a plurality of target genes related to protein expression,
wherein each primer set comprises a plurality of forward and reverse primer pairs specific to each target gene related to protein expression, wherein:
(i) each forward primer of the of the plurality of forward and reverse primer pairs specific to each target gene related to protein expression is complementary to a sequence spanning an exon-exon junction of each target gene related to protein expression,
wherein each reverse primer of the of the plurality of forward and reverse primer pairs specific to each target gene related to protein expression is complementary to a sequence about 100 base pairs downstream of the sequence spanning the exon-exon junction of each target gene related to protein expression,
wherein each reverse primer of the plurality of forward and reverse primer pairs specific to each target gene related to protein expression comprises a barcode sequence on its 5′ end, wherein the barcode sequence of each reverse primer corresponding to each target gene related to protein expression is different,
(ii) each reverse primer of the plurality of forward and reverse primer pairs specific to the plurality of target genes related to protein expression is complementary to a sequence spanning an exon-exon junction of each target gene related to protein expression,
wherein each forward primer of the plurality of forward and reverse primer pairs specific to the plurality of target genes related to protein expression is complementary to a sequence about 100 base pairs downstream of the sequence spanning the exon-exon junction of each target gene related to protein expression,
wherein each reverse primer of the plurality of forward and reverse primer pairs specific to the plurality of target genes related to protein expression comprises a barcode sequence on its 5′ end, wherein the barcode sequence of each reverse primer corresponding to each target gene related to protein expression is different; or
(iii) each forward and each reverse primer of the plurality of forward and reverse primer pairs specific to the plurality of target genes related to protein expression is complementary to consecutive sequences spanning an exon-exon junction of each target gene related to protein expression,
wherein each reverse primer of the plurality of forward and reverse primer pairs specific to the plurality of target genes related to protein expression comprises a barcode sequence on its 5′ end, wherein the barcode sequence of each reverse primer corresponding to each target gene related to protein expression is different,
thereby generating a plurality of amplicons;
(c) purifying the plurality of amplicons from step (b);
(d) amplifying the purified product from step (c) by using universal indexed adapter primers to generate a sequencing library;
(e) purifying the sequencing library obtained from step (d);
(f) subjecting the purified sequencing library from step (e) to multiplex sequencing on a next-generation sequencing platform to obtain a plurality of sequencing reads;
(g) deriving a consensus read of each sequence from the plurality of sequencing reads obtained from step (f);
(h) performing a sequence alignment of the consensus read obtained from step (g) to a reference genome,
(I) if the sequence alignment results in a partial alignment to the reference genome of an exon from a first gene and a partial alignment to the reference genome of an exon from a second gene, then:
(i) determining the sequence alignment as a split read,
(ii) counting/enumerating the number of split reads from step (h)(I)(i) that supports a fusion junction, and
(iii) if the number of split reads from step (h)(I)(ii) is two or more, then determining the first gene and the second gene as fusion partners,
(II) if the sequence alignment results in an alignment to the reference genome of the control housekeeping gene, then:
(i) determining the sequence alignment as a consensus read of the control housekeeping gene,
(ii) counting/enumerating consensus read pairs of the control housekeeping gene from step (h)(II)(i), and
(iii) determining the level of gene expression of the control housekeeping gene,
(III) if the sequence alignment results in an alignment to the reference genome of the target gene related to protein expression,
(i) determining the sequence alignment as a consensus read of the target gene related to protein expression,
(ii) counting/enumerating consensus read pairs of the target gene related to protein expression from step (h)(III)(i), and
(iii) determining the level of gene expression of the target gene related to protein expression;
(i) determining presence or absence of the genomic alteration and/or determining presence or absence of the gene expression and/or quantifying the level of the gene expression based on the sequence alignment from step (h).
|