| CPC G16B 40/00 (2019.02) [G16B 20/40 (2019.02)] | 48 Claims |
|
1. A method for configuring a machine learning model to model population frequency for variant classification, the method comprising:
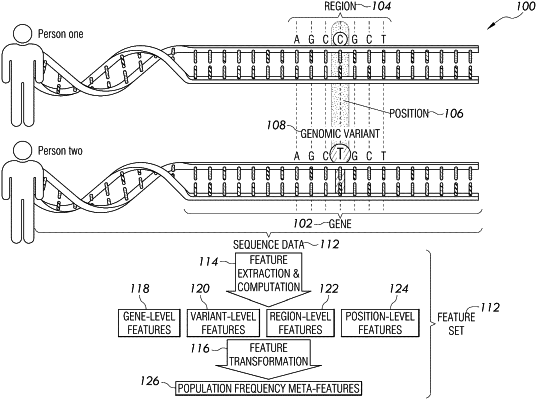
applying a logistic regression model to a first set of population data for a first set of genes, wherein an item of the first set of population data comprises, for a variant located at a position within a gene of the first set of genes, a set of features comprising at least one gene-level feature, at least one variant-level feature, and at least one population frequency meta-feature, and a reference label that indicates whether the variant is benign or pathogenic, wherein the at least one population frequency meta-feature quantifies predictive value of allele frequency in the gene, wherein the applying comprises computing a gene-level constraint; including the gene-level constraint in the at least one gene-level feature; computing an allele frequency; including the allele frequency in the at least one variant-level feature; including, in the at least one population frequency meta-feature, a mathematical combination of the gene-level constraint and the allele frequency;
and applying the logistic regression model to the set of features including the mathematical combination of the gene-level constraint and the allele frequency; wherein the trained logistic regression model is capable of outputting variant pathogenicity estimates that satisfy the at least one second performance criterion based on the set of features including the mathematical combination of the gene-level constraint and the allele frequency;
for each item of the first set of population data, evaluating a variant classification prediction output by the logistic regression model based on an expected variant classification indicated by the reference label; and
iteratively adjusting a value of at least one parameter or coefficient of the logistic regression model until output of a loss function computed based on the variant classification prediction output by the logistic regression model satisfies at least one first performance criterion, to produce a trained logistic regression model, wherein the trained logistic regression model is capable of outputting variant pathogenicity estimates that satisfy at least one second performance criterion.
|