| CPC G16B 20/00 (2019.02) [G06N 3/04 (2013.01); G06N 3/047 (2023.01); G06N 3/048 (2023.01); G06N 3/08 (2013.01); G06N 3/084 (2013.01); G16B 30/00 (2019.02); G16B 40/00 (2019.02); G16B 40/20 (2019.02); G16B 50/00 (2019.02); G06F 18/24 (2023.01)] | 20 Claims |
|
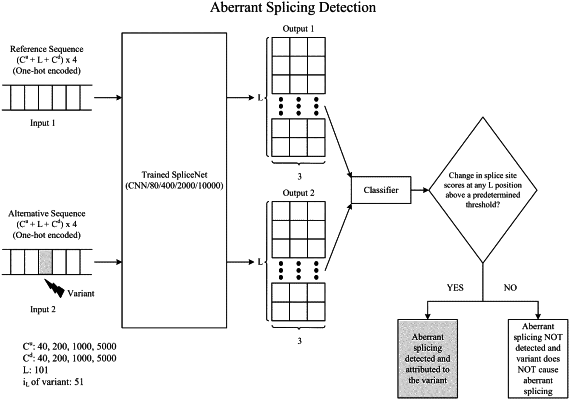
1. A system for aberrant splicing determination, including at least one processor coupled to memory, the memory loaded with instructions that, when executed by the at least one processor, cause the system to perform operations comprising:
a trained atrous convolutional neural network, running on the at least one processor, that processes pre-mRNA sequences, including:
an input layer that
receives a variant sequence with target nucleotides flanked on each side by flanking nucleotides; and
accesses a reference sequence corresponding to the variant sequence;
convolutional layers that perform atrous convolutions on the target nucleotides in the variant sequence and corresponding reference nucleotides in the reference sequence, and generate, for each of the target nucleotides and the corresponding reference nucleotides, a triplet splice site score comprising a donor site probability, an acceptor site probability, and a non-splicing site probability; and
an output layer that determines, from position-wise differences in respective triplet splice site scores of the target nucleotides and the corresponding reference nucleotides, whether a variant in the variant sequence causes aberrant splicing at any of the target nucleotides and is therefore pathogenic.
|