| CPC G01N 33/574 (2013.01) [G06F 18/211 (2023.01); G06T 7/0012 (2013.01); G06V 10/82 (2022.01); G06V 20/695 (2022.01); G06V 20/698 (2022.01); G16H 50/20 (2018.01); G06T 2207/10024 (2013.01); G06T 2207/20081 (2013.01); G06T 2207/20084 (2013.01); G06T 2207/30024 (2013.01)] | 21 Claims |
|
1. A method comprising:
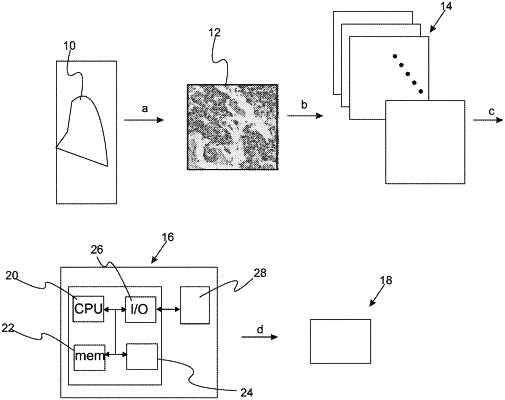
a) training an untrained machine learnable device to predict status of a diagnostic, prognostic, or theragnostic feature in a stained tissue sample, the untrained machine learnable device being a deep neural network which is trained with a characterized set of digital images of stained tissue samples, each digital image of the characterized set of digital images of stained tissue samples having a known status for the diagnostic, prognostic, or theragnostic feature and an associated 2-dimensional grid of spatial locations, wherein the known status for the diagnostic, prognostic, or theragnostic feature indicates presence or absence of a predetermined biomarker, wherein the predetermined biomarker comprises estrogen receptor; wherein training the untrained machine learnable device comprises:
identifying a plurality of extracted features in each digital image of the characterized set of digital images of stained tissue samples, wherein the extracted features comprise a nuclear morphometric feature comprising nuclear shape and/or nuclear orientation of the stained tissue samples;
associating a value for each extracted feature with each spatial location to form a set of extracted feature maps, each value being a single number, a vector, or a matrix, and each spatial location being defined by associated position coordinates, wherein each extracted feature map links values for extracted features to position coordinates of the associated 2-dimensional grid of spatial locations; and
inputting the set of extracted feature maps to the untrained machine learnable device to form associations therein between the set of extracted feature maps and the known status for the diagnostic, prognostic, or theragnostic feature, thereby creating a trained machine learnable device; and
b) predicting a status for the diagnostic, prognostic, or theragnostic feature of a stained tissue sample of unknown status for the diagnostic, prognostic, or theragnostic feature by:
obtaining a sample digital image for the stained tissue sample of unknown status, the sample digital image for the stained tissue sample of unknown status having an associated 2-dimensional grid of spatial locations;
associating a value for each extracted feature from the sample digital image for the stained tissue sample of unknown status with each spatial location of the sample digital image to form a test set of extracted feature maps for the stained tissue sample of unknown status; and
inputting the test set of extracted feature maps to the trained machine learnable device to obtain a predicted status for the status of the diagnostic, prognostic, or theragnostic feature for the stained tissue sample of unknown status, wherein a pre-processing step is performed in which classification results of a deep neural network trained on nuclear morphometric features is compared to a deep neural network trained on cytoplasmic and extracellular morphometric features to determine which morphometric features are predictive of an outcome.
|