| CPC G16H 50/70 (2018.01) [G06F 16/182 (2019.01); G06F 16/436 (2019.01); G16H 50/20 (2018.01); G06Q 50/22 (2013.01)] | 20 Claims |
|
1. A method comprising:
obtaining reads of biological samples of respective sample sources wherein each of the biological samples contains genomic material from a plurality of distinct microorganisms within an environment of a corresponding one of the sample sources; and
performing distributed data analytics to characterize an actual or potential outbreak of at least one of a disease, an infection and a contamination that involves genomic material from multiple ones of the distinct microorganisms in one or more of the sample sources;
wherein performing distributed data analytics comprises:
performing local analytics in respective ones of a plurality of data zones; and
performing global analytics utilizing results of the local analytics performed in the respective data zones;
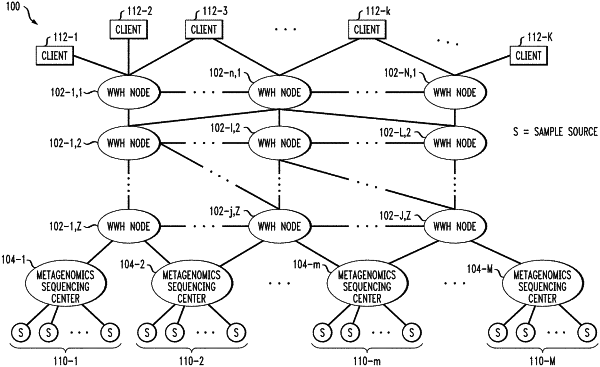
wherein each of the data zones comprises one or more sequencing centers utilized to generate a corresponding subset of the reads within that data zone;
wherein the local analytics performed in a given one of the data zones utilize reads of one or more of the biological samples sequenced in the one or more sequencing centers of the given data zone;
wherein the local analytics performed in the given data zone comprise analyzing the reads of the one or more biological samples against a local set of known gene units;
wherein at least one result of the global analytics comprises a graph in which nodes correspond to respective biological samples and edges between the nodes characterize epidemiological relationships between the biological samples, the edges being weighted by sample-to-sample comparison scores of metagenomics sequencing results for the biological samples; and
wherein the method is implemented by a distributed data processing system comprising a plurality of processing devices configured to communicate with one another over at least one network.
|