| CPC G16B 40/20 (2019.02) [C12Q 1/689 (2013.01); G16B 20/00 (2019.02); G16B 30/00 (2019.02); C12Q 1/6888 (2013.01); C12Q 2600/106 (2013.01); C12Q 2600/158 (2013.01)] | 20 Claims |
|
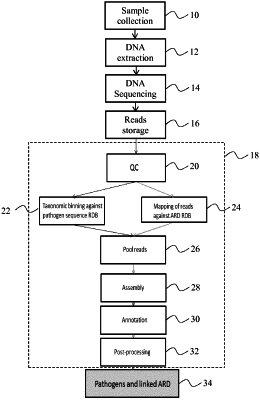
1. A method of analyzing a metagenomic sample, comprising:
processing the metagenomic sample to extract DNA from one or more pathogens if present in the metagenomic sample;
sequencing the extracted DNA to produce a set of digital nucleic acid sequences, i.e., “reads”, that have an average length of L bp in which L>100;
assigning a first subset of reads to a known pathogen by mapping the first subset of reads against genomic data of the known pathogen using a first database comprising genomic data for a plurality of known pathogens;
assigning a second subset of reads to a sequence of a known antibiotic resistance determinant or virulence genetic determinant of a length greater than that of the average length of the reads by mapping the second subset of reads against sequence data of the known antibiotic resistance determinant or virulence genetic determinant using a second database comprising sequence data for a plurality of antibiotic resistance determinants and/or virulence genetic determinants such that there are (i) reads within the second subset of reads that fall entirely within the sequence of the known antibiotic resistance determinant or virulence genetic determinant and (ii) reads within the second subset of reads that are astride the sequence of the known antibiotic resistance determinant or virulence genetic determinant;
producing a pool of reads comprising the first subset of reads and the second subset of reads; and
assembling the reads in the pool in order to produce at least one assembled digital nucleic acid, i.e., “contig”,
wherein the reads that are astride the sequence of the known antibiotic resistance determinant or virulence genetic determinant each have (i) a first portion falling inside of the sequence and (ii) a second portion falling outside of the sequence in the range [1; L-50] bp.
|