| CPC G06T 7/0012 (2013.01) [G06V 10/462 (2022.01); G06V 10/50 (2022.01); G06V 10/52 (2022.01); G06V 10/761 (2022.01); G06V 20/695 (2022.01); G06V 20/698 (2022.01); G06T 2207/30056 (2013.01); G06T 2207/30096 (2013.01)] | 6 Claims |
|
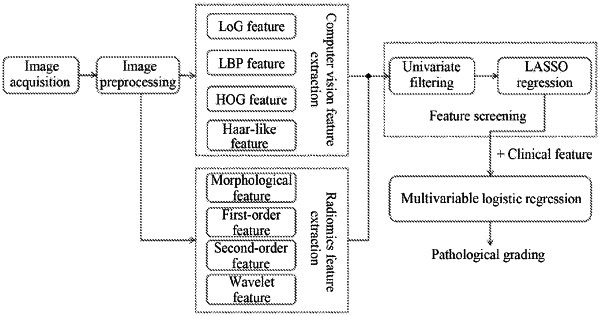
1. A method for classifying hepatocellular carcinoma images by combining computer vision features and radiomics features, comprising:
step (1), obtaining and preprocessing a hepatocellular carcinoma image, wherein the preprocessing comprises: segmenting a hepatic tumor region in the hepatocellular carcinoma image, re-sampling volume pixels of a segmented image of the hepatic tumor region into a space of a fixed size, normalizing a grey scale thereof, and labeling a corresponding pathological grading result;
step (2), extracting computer vision features from the segmented image of the hepatic tumor region, wherein the computer vision features comprise Location of a Gaussian (LoG) filter features, Local Binary Patterns (LBP) features, Histogram of Oriented Gradients (HOG) features, and haar-like features;
step (3), extracting radiomics features from the segmented image of the hepatic tumor region, wherein the radiomics features comprise morphological features, grey scale features, texture features, and wavelet features;
step (4), jointing the computer vision features extracted in step (2) and the radiomics features extracted in step (3) together, and screening features in a merged feature set X={X1, X2, . . . , Xi, . . . , Xn} resulted from the joint, wherein n represents a number of feature vectors in the merged feature set, Xi represents an ith feature vector in the merged feature set Xi={xi1, xi2, . . . , xim}, Xi represents an ith element in Xi, and m represents a number of elements in Xi; wherein the screening features in the merged feature set comprises:
step (4.1), firstly, excluding single-valued features, features having variances less than a preset threshold, features irrelevant to the corresponding pathological grading, and redundant features by univariate filtering;
wherein a relationship intensity between features and the corresponding pathological grading is measured through mutual information calculations, so that the features having mutual information values lower than the preset threshold are referred to as irrelevant to the pathological grading, and the mutual information calculations are performed according to:
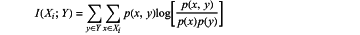 wherein, Xi represents an ith feature vector in the merged feature set, Y represents a label, p(x, y) is a joint probability density function of X and Y, and p(x) and p(y) are, respectively, marginal probability density functions of X and Y;
the redundant features are those having correlation coefficients greater than 0.9, and a correlation coefficient is to be computed according to:
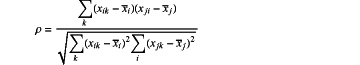 wherein, ρ is a calculated result of the correlation coefficient, xik represents a kth element in the ith feature vector in the merged feature set, xjk represents a kth element in a jth feature vector in the merged feature set, and xi and xj respectively represent mean values of all elements in an ith feature vector and a jth feature vector in the merged feature set;
step (4.2), adopting a feature dimension reduction of a Least Absolute Shrinkage and Selection Operator (Lasso) regression model to train those unhelpful feature parameters to be 0 in order to obtain a sparse solution; wherein a cost function of the Lasso regression is:
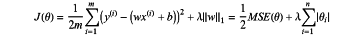 Wherein, m is a number of samples, n is a number of features, x(i) represents all feature values of an ith sample, y(i) represents a label value of the jth sample, w is a parameter vector of a length n, ∥w1∥ represents an L1 norm of the parameter vector w, b represents a constant coefficient, and λ is a parameter for controlling a regression complexity, wherein a best value of λ is selected by taking a binomial deviation as a criterion and then iterated by a 10-fold cross validation method to apply a λ value of the binomial deviation in the Lasso regression model; and a predict score is obtained by multiplying Lasso-selected features by their respective coefficients to provide products and then summing the products;
step (5), after clinical features are obtained from a hepatic tumor region image and all the clinical features are deleted one by one, adopting a multivariable logistic regression, by combining the image with all feature matrices resulted from screening in step (4), to get a classifier for pathological grading of hepatocellular carcinoma for different combinations; using an Akaike information criterion (AIC) as a criterion to search backward a most proper combination thereof, and then according to an influence of the binomial deviation and the number of variables during selection selecting a model of a lowest AIC score as a final classifier for pathological grading of hepatocellular carcinoma which may input a best combined feature matrix; wherein the AIC is defined as:
AIC=−2 ln(L)+2k
wherein L is a maximum likelihood under this model, and k is a number of model variables;
step (6), acquiring the hepatocellular carcinoma image to go through the pathological grading of hepatocellular carcinoma and processing according to step (1) to segment an image of the hepatic tumor region from which features of a same type as that of the best combined feature matrix said in step (5) are extracted as a feature matrix to be graded; and inputting the feature matrix to be graded into the classifier for pathological grading of hepatocellular carcinoma obtained in step (5) to output a pathological grading result of hepatocellular carcinoma.
|