| CPC C12Q 1/6874 (2013.01) [C12Q 1/6806 (2013.01); C12Q 1/6869 (2013.01)] | 30 Claims |
|
1. A method for identifying variant nucleotides in a population of cell-free nucleic acids comprising:
(a) contacting a population of cell-free nucleic acids comprising double-stranded DNA molecules with single-stranded overhangs at one or both ends with a protein having 5′-3′ polymerase activity and a 3′-5′ exonuclease activity, wherein the protein digests 3′ overhangs and fills in 5′ overhangs with complementary nucleotides, to generate double-stranded DNA molecules with one or both ends blunt;
(b) tailing blunt ends of the DNA molecules and ligating the resulting DNA molecules to one or more adapters with a complementary tail;
(c) determining sequences of a plurality of the double-stranded DNA molecules to provide sequenced DNA molecules;
(d) for each designated position in a reference sequence,
(i) identifying a subset of sequenced DNA molecules including the designated position, and
(ii) identifying sequenced DNA molecules in the subset in which the designated position is occupied by a variant nucleotide; and
(e) calling presence of a variant nucleotide at each designated position for which the sequenced DNA molecules in step (d)(ii) support the call, except that presence of a variant nucleotide at a designated position is not called if:
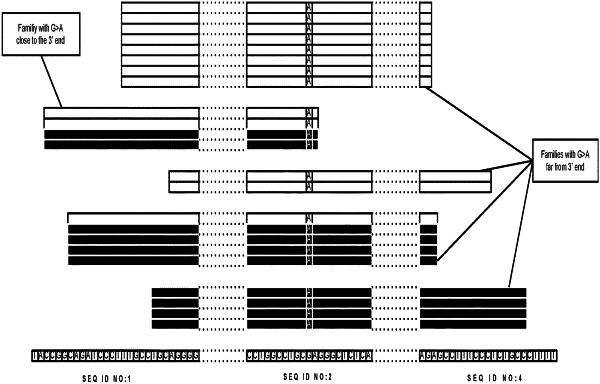
(i) the variant is a C to T or G to A variation compared with the reference nucleotide; and
(ii) the variant nucleotide is classified as a deamination error based on:
(1) nucleotide context around the designated position and/or
(2) distance of the C to T variation at the designated position from the 5′-end in sequenced DNA molecules in the subset or distance of the G to A variation at the designated position from the 3′-end in sequenced DNA molecules in the subset, wherein at least one variant nucleotide which would otherwise have been called in step (e) is not called due to conditions (i) and (ii) being determined to have been met.
|