| CPC G16B 40/10 (2019.02) [G01N 33/6818 (2013.01); G01N 33/6848 (2013.01); G06N 3/02 (2013.01); G16B 30/20 (2019.02); G16B 40/20 (2019.02); H01J 49/0036 (2013.01)] | 36 Claims |
|
1. A computer implemented system for de novo sequencing of a peptide from mass spectrometry data acquired by data-independent acquisition using neural networks, the computer implemented system comprising:
at least one memory and at least one processor configured to receive:
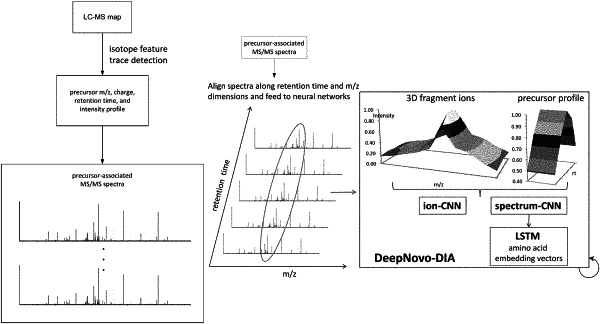
a first input representing at least one precursor profile, each precursor profile representing intensities of one or more precursor ion signals associated with a precursor retention time;
a second input representing a plurality of fragment ion spectra for each precursor profile, each fragment ion spectra representing:
signals from fragment ions generated from an associated precursor ion, and
a fragment retention time; and
provide a plurality of layered computing nodes configured to form an artificial neural network for generating a probability measure for one or more candidates to a next amino acid in an amino acid sequence, the artificial neural network trained on mass spectrometry data containing retention time, a plurality of fragment ions peaks of sequences differing in length and differing by one or more amino acids;
wherein the plurality of layered nodes are configured to receive a mass spectrometry spectrum data base on the first and second inputs, the mass spectrometry spectrum data representing the at least one precursor profile and the fragment ion spectra, the plurality of layered nodes comprising at least one convolutional layer for filtering mass spectrometry spectrum data to detect fragment ion peaks; and
wherein the processor is configured to:
receive an input prefix representing a determined amino acid sequence of the peptide,
provide the mass spectrometry spectrum data to the plurality of layered nodes,
identify a next amino acid based on a candidate next amino acid having a greatest probability measure based on the output of the artificial neural network and the mass spectrometry spectrum data of the peptide;
update the determined amino acid sequence with the next amino acid,
and generate an output signal representing a final determined sequence;
wherein the plurality of layered nodes receives a matrix data representing the mass spectrometry spectrum data, and output a probability measure vector; and wherein the second input comprises a matrix data representing:
i. batch size,
ii. number of amino acids;
iii. ion types;
iv. number of fragment ion spectra associated with a precursor profile; and
v. window size for filtering fragment ion peaks.
|