| CPC G06F 16/24558 (2019.01) [G06F 16/2246 (2019.01); G06F 21/60 (2013.01)] | 7 Claims |
|
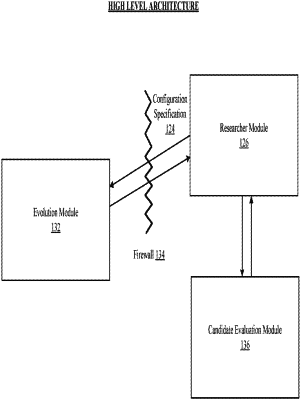
1. A method for facilitating evolutionary optimization of candidate genomes in a desired domain, wherein evolution of the candidate genomes and evaluation of the candidate genomes with domain specific data are performed securely from each other, the method:
receiving by an evolution module located in a first secure processing environment from a researcher module a configuration specification, wherein the researcher module automatically builds the configuration specification using defined rules;
parsing the configuration specification by the evolution module into a tree structure of specific coded data structures;
generating a policy assembly using the parsed tree structure of specific coded data structures to produce a representation tree of reproduction policy structures, wherein each instance of the reproduction policy structures represents one of a single leaf node or branch node in the representation tree;
creating by the evolution module an internal representation of a candidate genome in accordance with the representation tree of reproduction policy structures, wherein the reproduction policy structures are data-type specific;
translating by the evolution module the internal representation of the candidate genome to a known representation of the candidate genome, wherein the known representation of the candidate genome is known to a candidate evaluation module; and
providing the known representation of the candidate genome to the candidate evaluation module located in a second secure processing environment for fitness evaluation in the desired domain.
|