| CPC G16B 20/20 (2019.02) [G16B 20/10 (2019.02); G16B 40/00 (2019.02); G16B 40/30 (2019.02); G16B 20/00 (2019.02)] | 17 Claims |
|
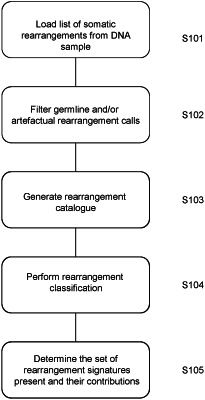
1. A method of detecting rearrangement signatures in a previously obtained DNA sample from a patient, the method comprising:
obtaining whole genome DNA sequence data from said DNA sample by sequencing nucleic acid material obtained from the DNA sample, wherein the DNA sample comprises fresh-frozen derived DNA, circulating tumor DNA, or formalin-fixed paraffin-embedded (FFPE) DNA representative of a suspected or known tumor from the patient;
obtaining, by a processor, a file of somatic mutations in the DNA sample, the somatic mutations obtained from the whole genome DNA sequence data;
filtering, by the processor and using data obtained from one or more databases, the somatic mutations in the file to remove one or more germline rearrangements and copy number polymorphisms;
filtering, by the processor and using data in alignment mapping files, the somatic mutations in the file to remove known sequencing artifacts;
cataloguing, by the processor, the somatic mutations in the file to produce a rearrangement catalogue for the DNA sample which classifies rearrangement mutations identified in the whole genome DNA sequence data into a plurality of categories, the cataloguing comprising:
identifying mutations as one of: tandem duplications, deletions, inversions or translocations and grouping mutations identified as tandem duplications, deletions or inversions by size, and
identifying mutations as being clustered or non-clustered;
determining, by said processor, the contributions of known rearrangement signatures to said rearrangement catalogue using a measure of similarity between the rearrangement mutations in said rearrangement catalogue and the known rearrangement signatures, wherein each of the known rearrangement signatures comprises proportions of rearrangements in the plurality of categories and wherein said known rearrangement signatures are indicative of the presence of mutational processes contributing to cancer; and
determining, by the processor, if the number or proportion of rearrangements in the rearrangement catalogue which are determined to be associated with a particular one of the known rearrangement signatures exceeds a predetermined threshold and, if so, determining that the particular one rearrangement signature is present in the DNA sample.
|