| CPC G01N 33/6818 (2013.01) [G01N 33/582 (2013.01); G01N 33/6812 (2013.01); G01N 2333/95 (2013.01)] | 19 Claims |
|
1. A method for identifying a protein in a sample, comprising:
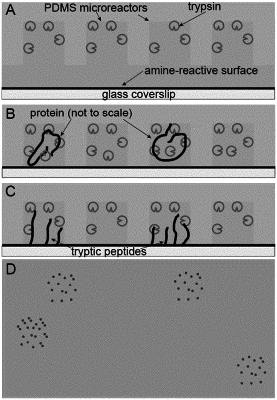
A. introducing at least one protein molecule onto a bead, and wherein the bead contains an amine-reactive surface and wherein the amine-reactive surface allows for direct reaction between the surface and the N-termini of a peptide and the protein molecule has been treated such that the N-terminals are capable of reacting to the amine-reactive surface;
B. incubating an endopeptidase with the at least one protein molecule on the bead for a time and under conditions which allow the endopeptidase to cleave the protein into peptides, wherein the endopeptidase cleaves the at least one protein molecule at specific amino acids in the protein resulting in peptides comprising an N-terminal amino acid and corresponding to the at least one protein molecule contained on the bead;
C. allowing the peptides to react via their N-terminal amino acid residue with the amine-reactive surface of the bead, for a time and under conditions which result in the peptides attaching to the amine-reactive surface of the bead and forming a pattern on the amine-reactive surface, wherein the pattern is associated with the bead containing the at least one protein molecule;
D. washing the endopeptidase from the bead, wherein the washing of the endopeptidase ends the cleaving of the protein in step B;
E. labeling the peptides at specific amino acid residues, such that the specific amino acid residues can be detected when imaged;
F. imaging the peptides to detect the labeled amino acids and generating amino acid sequence information from the peptides in step B;
G. further incubating the peptides generated in step B, with at least one enzyme or chemical for a time and under conditions which allow the enzyme or chemical to cleave and imaging the resulting peptides to generate additional amino acid sequence information;
H. washing the enzyme or chemical from the bead, wherein the washing of the enzyme or chemical ends the cleaving of the peptides in step G;
I. further labeling the peptides generated in step G, at specific amino acids different from the label in step E:
J. imaging the peptides to detect the labeled amino acids and generating additional amino acid sequence information; K. compiling the generated amino acid sequence information from steps F, G and J to obtain a putative amino acid sequence of the at least one protein molecule;
L. comparing the putative amino acid information of at least one protein molecule with a known amino acid sequence of proteins; and
M. identifying the protein from the comparison in step L; wherein steps G.-J. are repeated with a plurality of enzymes or chemical and labels as desired until a putative amino acid sequence of the at least one protein molecule is obtained.
|