| CPC G16B 10/00 (2019.02) [G06F 16/2228 (2019.01); G16B 30/10 (2019.02); G16B 40/20 (2019.02)] | 15 Claims |
|
1. A processor-implemented method for profiling of metagenome sample comprising:
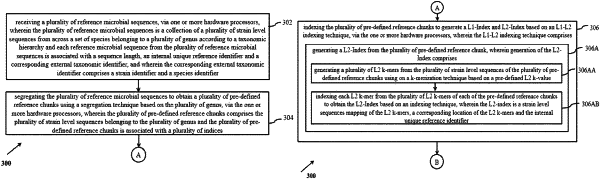
receiving a plurality of reference microbial sequences, via one or more hardware processors, wherein the plurality of reference microbial sequences is a collection of a plurality of strain level sequences from across a set of species belonging to a plurality of genus according to a taxonomic hierarchy and each reference microbial sequence from the plurality of reference microbial sequences is associated with a sequence length, an internal unique reference identifier and a corresponding external taxonomic identifier, and wherein the corresponding external taxonomic identifier comprises a strain identifier and a species identifier;
segregating the plurality of reference microbial sequences to obtain a plurality of pre-defined reference chunks using a segregation technique based on the plurality of genus, via the one or more hardware processors, wherein the plurality of pre-defined reference chunks comprises the plurality of strain level sequences belonging to the plurality of genus and the plurality of pre-defined reference chunks is associated with a plurality of indices;
indexing the plurality of pre-defined reference chunks to generate a L1-Index and L2-Index based on an L1-L2 indexing technique, via the one or more hardware processors, wherein the L1-L2 indexing technique comprises:
generating a L2-Index from the plurality of pre-defined reference chunks, wherein generation of the L2-Index comprises:
generating a plurality of L2 k-mers from the plurality of strain level sequences of the plurality of pre-defined reference chunks using on a k-merization technique based on a pre-defined L2 k-value;
indexing each L2 k-mer from the plurality of L2 k-mers of each of the pre-defined reference chunks to obtain the L2-Index based on an indexing technique, wherein the L2-index is a strain level sequences mapping of the L2 k-mers, a corresponding location of the L2 k-mers and the internal unique reference identifier;
generating a L1-Index from the plurality of pre-defined reference chunks, wherein generation of the L1-Index comprises:
generating a plurality of L1 k-mers from the plurality of strain level sequences of the plurality of pre-defined reference chunks using on the k-merization technique based on a pre-defined L1 k-value;
indexing the plurality of L1 k-mers to obtain the L1-Index based on a binary encoding technique and a numerical encoding technique, wherein the L1-Index is a map of the L1 k-mers and the indices of the plurality of pre-defined reference chunks;
receiving a plurality of microbial read sequences, via one or more hardware processors, wherein the microbial read sequences is a metagenome sample and each microbial read sequence from the plurality of microbial read sequences is associated with a read length and a sequencing depth; and
profiling the plurality of microbial read sequences, via one or more hardware processors, based on the L1-Index and the L2-Index, wherein the profiling the plurality of microbial read sequences at a profile taxonomic hierarchy comprises:
generating a plurality of L1 query k-mers and a plurality of L2 query k-mers for the plurality of microbial read sequences based on a pre-defined L1 k-value and a pre-defined L2 k-value respectively using the k-merization technique;
identifying from the L1-index, a matching reference plurality of pre-defined reference chunks and the corresponding plurality of indices for the plurality of L1 query k-mers;
identifying from the L2-Index, the plurality of strain level sequences and the internal unique reference identifier for the plurality of L2 query k-mers based on the matching reference plurality of pre-defined reference chunks and the plurality of indices; and
profiling the plurality of microbial read sequences, wherein the profiling comprises performing an abundance estimation of the plurality of strain level sequences based on an estimation technique using the strain level sequences, the internal unique reference identifier and the sequence length.
|